m (→Division Control) |
(→Lab-on-chip systems) |
||
| (41 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| − | The development of biological devices that transform living cells into biosensors, nano-factories, and therapeutic agents is the objective of synthetic biology. | + | The development of biological devices that transform living cells into biosensors, nano-factories, and therapeutic agents is the objective of synthetic biology. Georgiev Lab is a university wide research laboratory interested in innovations that make such devices reliable and efficient enough to make the biological systems a generally useful technology. We apply tools from engineering and synthetic biology to 1) build electromechanical devices co-designed with biological devices, 2) derive estimation and modeling methods specifically for biological systems, and 3) develop biological control mechanisms and theory. |
If you are interested in joining us in this effort or if you are just simply interested, contact Dr. Daniel Georgiev (georgiev@kky.zcu.cz). | If you are interested in joining us in this effort or if you are just simply interested, contact Dr. Daniel Georgiev (georgiev@kky.zcu.cz). | ||
| − | == | + | ==[[Projects/Synthetic Biology|Synthetic Biology]]== |
| − | [[File: | + | [[File:New_lab.jpg|thumb|right|alt=-|Synthetic Biology wet lab.]] |
| − | + | Synthetic Biology is often misinterpreted to be a discipline encompassing the available biological (cloning, cultivation, analysis, etc.) protocols as well as the established application areas (bioproduction, adaptive therapy, etc.). While the laboratory limitations and application needs are important inputs and outputs to this area, we believe the primary and direct contribution of Synthetic biology is a systematic engineering method for developing biotechnology. Our research focuses on practical design rules and efficient tuning algorithms with the purpose of making the synthesis of regulatory and signalling devices faster and more reliable. | |
| − | + | <br> | |
| − | == | + | ==Bioinformatics== |
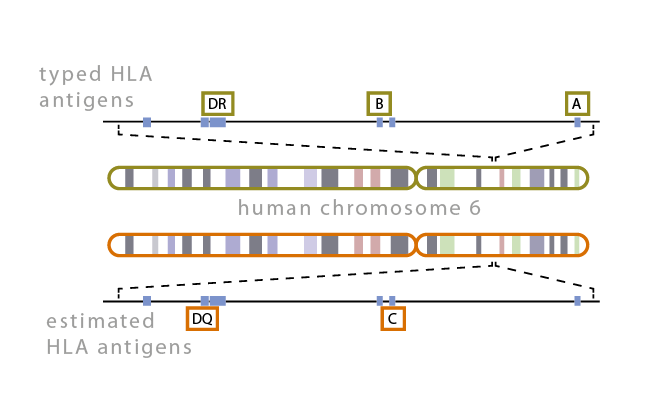
| − | [[File: | + | [[File:HLACompatibility.png|thumb|right|alt=HLA compatibility figure.|HLA compatibility.]] |
| − | + | Compatibility of the host and donor human leukocyte antigen systems is critical to the success of a transplant. Discrepancies in HLA loci produce host immune responses that lead to graft rejection. Full donor DNA sequencing, however, is too costly and hence partial typization of important loci is carried out. Georgiev Lab in collaboration with the University Hospital in Pilsen is interested in developing computational tools for the estimation of the missing DNA data. | |
| + | * [http://ccy.zcu.cz/registr/CNRDDstatistika.html MADORA] - Marrow Donor Register Analytics (MADORA) for the Czech National Register of Marrow Donors (CNRDD) | ||
| + | * [http://ccy.zcu.cz/registr/CZ-HANA.html HANA] - Haplotype analyzer (HANA) for the Czech population. | ||
| + | <br> | ||
| − | == | + | ==[[Projects/Microfluidics|Lab-on-chip systems]]== |
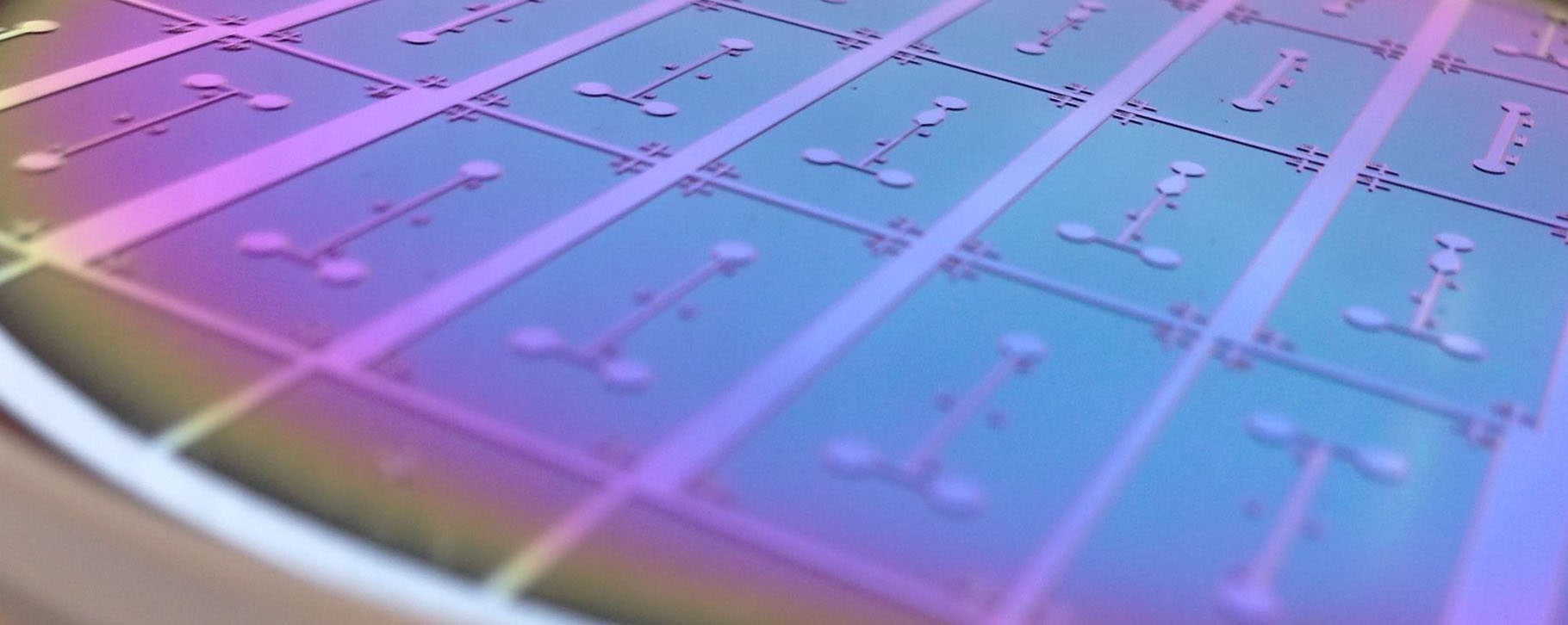
| − | [[File: | + | [[File:master.jpg|thumb|right|alt=Silicon master for the PDMS soft-lithography.|A soft-lithography silicon master used for fabrication of the microfluidic devices at the Georgiev Lab.]] |
| − | + | ||
| − | + | Lab-on-chip systems integrate one or more lab functions on a chip of just couple centimeters squared. Such systems are able to work with extremely low volumes of liquid samples, can significantly decrease costs of individual operations and decrease requirements on additional hardware. Hence they enable for instance implementation of affordable diagnostic tools for cancer or development of tools enabling selection of optimal therapy increasing treatment efficiency and its personalization for individual patients. | |
| + | Our research focuses on applications of the electric field in lab-on-chip systems in the form of dielectrophoretic effect based on polarization effect in the electric field. Dielectrophoretic effect may be used for measurement of cell properties on population level, their selective contactless manipulation or immobilization. At the same time, it may be combined with other electric field-based methods such as the electro-rotation, the impedance spectroscopy and the electro-poration, and with other technologies such as microscopy and optical data analysis. | ||
| − | < | + | <br> |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
Latest revision as of 09:09, 12 May 2017
The development of biological devices that transform living cells into biosensors, nano-factories, and therapeutic agents is the objective of synthetic biology. Georgiev Lab is a university wide research laboratory interested in innovations that make such devices reliable and efficient enough to make the biological systems a generally useful technology. We apply tools from engineering and synthetic biology to 1) build electromechanical devices co-designed with biological devices, 2) derive estimation and modeling methods specifically for biological systems, and 3) develop biological control mechanisms and theory.
If you are interested in joining us in this effort or if you are just simply interested, contact Dr. Daniel Georgiev (georgiev@kky.zcu.cz).
Synthetic Biology
Synthetic Biology is often misinterpreted to be a discipline encompassing the available biological (cloning, cultivation, analysis, etc.) protocols as well as the established application areas (bioproduction, adaptive therapy, etc.). While the laboratory limitations and application needs are important inputs and outputs to this area, we believe the primary and direct contribution of Synthetic biology is a systematic engineering method for developing biotechnology. Our research focuses on practical design rules and efficient tuning algorithms with the purpose of making the synthesis of regulatory and signalling devices faster and more reliable.
Bioinformatics
Compatibility of the host and donor human leukocyte antigen systems is critical to the success of a transplant. Discrepancies in HLA loci produce host immune responses that lead to graft rejection. Full donor DNA sequencing, however, is too costly and hence partial typization of important loci is carried out. Georgiev Lab in collaboration with the University Hospital in Pilsen is interested in developing computational tools for the estimation of the missing DNA data.
- MADORA - Marrow Donor Register Analytics (MADORA) for the Czech National Register of Marrow Donors (CNRDD)
- HANA - Haplotype analyzer (HANA) for the Czech population.
Lab-on-chip systems
Lab-on-chip systems integrate one or more lab functions on a chip of just couple centimeters squared. Such systems are able to work with extremely low volumes of liquid samples, can significantly decrease costs of individual operations and decrease requirements on additional hardware. Hence they enable for instance implementation of affordable diagnostic tools for cancer or development of tools enabling selection of optimal therapy increasing treatment efficiency and its personalization for individual patients. Our research focuses on applications of the electric field in lab-on-chip systems in the form of dielectrophoretic effect based on polarization effect in the electric field. Dielectrophoretic effect may be used for measurement of cell properties on population level, their selective contactless manipulation or immobilization. At the same time, it may be combined with other electric field-based methods such as the electro-rotation, the impedance spectroscopy and the electro-poration, and with other technologies such as microscopy and optical data analysis.