(→Gibson assembly) |
|||
| (One intermediate revision by the same user not shown) | |||
| Line 8: | Line 8: | ||
[[File:Polymerase_fidelity.jpg|600px|center|Polymerase fidelity comparison]] | [[File:Polymerase_fidelity.jpg|600px|center|Polymerase fidelity comparison]] | ||
* Hairpins: It is formed by intramolecular interaction within the primer and should be avoided. Optimally a 3' end hairpin with a ΔG of -2 kcal/mol and an internal hairpin with a ΔG of -3 kcal/mol is tolerated generally - see [http://www.premierbiosoft.com/tech_notes/PCR_Primer_Design.html Primer Design Guide for PCR] | * Hairpins: It is formed by intramolecular interaction within the primer and should be avoided. Optimally a 3' end hairpin with a ΔG of -2 kcal/mol and an internal hairpin with a ΔG of -3 kcal/mol is tolerated generally - see [http://www.premierbiosoft.com/tech_notes/PCR_Primer_Design.html Primer Design Guide for PCR] | ||
| + | |||
| + | |||
| + | === Gibson assembly === | ||
| + | |||
| + | ==== Overhangs design ==== | ||
| + | * Tm of every overhang should be between 51-55°C (using Primer3) | ||
| + | * No hairpin should be present in the overhang (Primer3) | ||
| + | * No restriction sites and palindromes >5bp should be present (if you need to introduce a restriction site, put it between the annealing and the overlapping part of the primer) | ||
| + | * GC content should be kept around 50% | ||
| + | * Number of G or C in a row must be less than 5 | ||
| + | * Using NUPACK, check the secondary structure of fragment's end (i.e. overhang with a part of the fragment, about 100bp long) | ||
| + | |||
| + | ==== Reaction ==== | ||
| + | * Use the official [https://www.neb.com/protocols/2012/12/11/gibson-assembly-protocol-e5510 NEB manual] | ||
| + | * Use 50ng of vector | ||
| + | * Incubation time should be 2h | ||
| + | * In case of assembling parts without a vector, use equimolar ratios (otherwise, use 4x more inserts) | ||
Latest revision as of 16:38, 3 May 2015
Experiences
PCR
- With your low GC, you should also consider lowering your extension (not the annealing) temperature and doubling the extension time. This will allow you to PCR very low GC regions, which otherwise cannot be extended.
- When adding restriction sites to your primers, check that there is sufficient space if that site is near the end of the primer for the enzyme to be able to sufficiently bind and cut. As a general rule, 4-6 bp is a good length to protect your site with although this can vary to be as few as 1bp and as many as 9bp. For more information, visit the NEB information site.
- The fact that primers are synthesized 3' to 5' means that all or nearly all primers are correct and identical at the 3' end, which is important for locating the site of biological synthesis. Synthesized primers typically have a significant error rate, with most 100 bp primers being incorrect in one or more locations. The most common error is a truncation of the 5' end, followed by point deletions of single bases. Long strings of G's are problematic for synthesis.
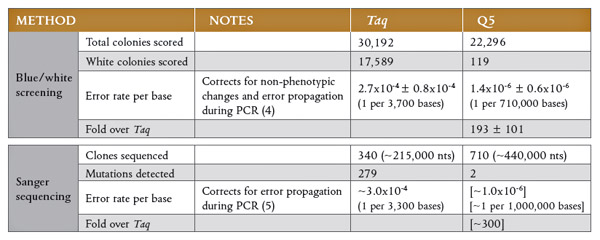
- Always choose a proper polymerase for your application:
- Hairpins: It is formed by intramolecular interaction within the primer and should be avoided. Optimally a 3' end hairpin with a ΔG of -2 kcal/mol and an internal hairpin with a ΔG of -3 kcal/mol is tolerated generally - see Primer Design Guide for PCR
Gibson assembly
Overhangs design
- Tm of every overhang should be between 51-55°C (using Primer3)
- No hairpin should be present in the overhang (Primer3)
- No restriction sites and palindromes >5bp should be present (if you need to introduce a restriction site, put it between the annealing and the overlapping part of the primer)
- GC content should be kept around 50%
- Number of G or C in a row must be less than 5
- Using NUPACK, check the secondary structure of fragment's end (i.e. overhang with a part of the fragment, about 100bp long)
Reaction
- Use the official NEB manual
- Use 50ng of vector
- Incubation time should be 2h
- In case of assembling parts without a vector, use equimolar ratios (otherwise, use 4x more inserts)