(→Microfluidics Research Laboratory) |
|||
| Line 3: | Line 3: | ||
If you are interested in joining us in this effort or if you are just simply interested, contact Dr. Daniel Georgiev (georgiev@kky.zcu.cz). | If you are interested in joining us in this effort or if you are just simply interested, contact Dr. Daniel Georgiev (georgiev@kky.zcu.cz). | ||
| − | == | + | ==Bioinformatics== |
[[File:HLACompatibility.png|thumb|right|alt=HLA compatibility figure.|HLA compatibility.]] | [[File:HLACompatibility.png|thumb|right|alt=HLA compatibility figure.|HLA compatibility.]] | ||
Compatibility of the host and donor human leukocyte antigen systems is critical to the success of a transplant. Discrepancies in HLA loci produce host immune responses that lead to graft rejection. Full donor DNA sequencing, however, is too costly and hence partial typization of important loci is carried out. Georgiev Lab in collaboration with the University Hospital in Pilsen is interested in developing computational tools for the estimation of the missing DNA data. | Compatibility of the host and donor human leukocyte antigen systems is critical to the success of a transplant. Discrepancies in HLA loci produce host immune responses that lead to graft rejection. Full donor DNA sequencing, however, is too costly and hence partial typization of important loci is carried out. Georgiev Lab in collaboration with the University Hospital in Pilsen is interested in developing computational tools for the estimation of the missing DNA data. | ||
| − | == | + | ==Synthetic Biology== |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
[[File:wiki_GeneTuning.png|thumb|right|alt=gfp under control of placI.|GFP under control of pLacI.]] | [[File:wiki_GeneTuning.png|thumb|right|alt=gfp under control of placI.|GFP under control of pLacI.]] | ||
Transcription networks are commonly used in synthetic biology to implement a wide variety of regulatory, logic, and temporal functions. Tuning of transcription networks is commonly achieved by design of the gene promoter regions. Characterized promoter libraries serve well in this design as an initial starting point (Hammer 2006). As with any engineered system, however, precise behavior is attained by in vivo fine tuning. Model based tuning is made difficult by inherent biological model overparametrization (Gutenkunst 2008). Tuning using high throughput assays is also prohibitive both in terms of experimental workload and precision. Georgiev Lab is working on developing precise model-free tuning protocols. These are protocols that are able to identify small differences between competing promoter designs and isolate the ones that yield desirable behaviors, e.g., robustness to common perturbations, fast activation times, sufficient temporal spacings, and correct equilibrium concentrations. | Transcription networks are commonly used in synthetic biology to implement a wide variety of regulatory, logic, and temporal functions. Tuning of transcription networks is commonly achieved by design of the gene promoter regions. Characterized promoter libraries serve well in this design as an initial starting point (Hammer 2006). As with any engineered system, however, precise behavior is attained by in vivo fine tuning. Model based tuning is made difficult by inherent biological model overparametrization (Gutenkunst 2008). Tuning using high throughput assays is also prohibitive both in terms of experimental workload and precision. Georgiev Lab is working on developing precise model-free tuning protocols. These are protocols that are able to identify small differences between competing promoter designs and isolate the ones that yield desirable behaviors, e.g., robustness to common perturbations, fast activation times, sufficient temporal spacings, and correct equilibrium concentrations. | ||
| Line 24: | Line 19: | ||
<mediaplayer width='550' height='300'>http://www.ccy.zcu.cz/movies/Osc_video.mov</mediaplayer> | <mediaplayer width='550' height='300'>http://www.ccy.zcu.cz/movies/Osc_video.mov</mediaplayer> | ||
| − | ==Microfluidics | + | ==[[Projects/Microfluidics|Microfluidics]]== |
[[File:master.jpg|thumb|right|alt=Silicon master for the PDMS soft-lithography.|A soft-lithography silicon master used for fabrication of the microfluidic devices at the Georgiev Lab.]] | [[File:master.jpg|thumb|right|alt=Silicon master for the PDMS soft-lithography.|A soft-lithography silicon master used for fabrication of the microfluidic devices at the Georgiev Lab.]] | ||
| − | Our | + | Our Microfluidics Research focuses on the study of microfluidic transport phenomena and the design of microfluidic devices with applications in synthetic biology. Current research projects include design and development of sensitive cell sorting techniques, on-chip cell culture, bio-sensors and cell-to-cell communication analysis. |
Revision as of 15:17, 5 March 2015
The development of biological devices that transform living cells into biosensors, nano-factories, and therapeutic agents is the objective of synthetic biology. Georgiev Lab is a university wide research laboratory interested in innovations that make such devices reliable and efficient enough to make the biological systems a generally useful technology. We apply tools from engineering and synthetic biology to 1) build electromechanical devices co-designed with biological devices, 2) derive estimation and modeling methods specifically for biological systems, and 3) develop biological control mechanisms and theory.
If you are interested in joining us in this effort or if you are just simply interested, contact Dr. Daniel Georgiev (georgiev@kky.zcu.cz).
Bioinformatics
Compatibility of the host and donor human leukocyte antigen systems is critical to the success of a transplant. Discrepancies in HLA loci produce host immune responses that lead to graft rejection. Full donor DNA sequencing, however, is too costly and hence partial typization of important loci is carried out. Georgiev Lab in collaboration with the University Hospital in Pilsen is interested in developing computational tools for the estimation of the missing DNA data.
Synthetic Biology
Transcription networks are commonly used in synthetic biology to implement a wide variety of regulatory, logic, and temporal functions. Tuning of transcription networks is commonly achieved by design of the gene promoter regions. Characterized promoter libraries serve well in this design as an initial starting point (Hammer 2006). As with any engineered system, however, precise behavior is attained by in vivo fine tuning. Model based tuning is made difficult by inherent biological model overparametrization (Gutenkunst 2008). Tuning using high throughput assays is also prohibitive both in terms of experimental workload and precision. Georgiev Lab is working on developing precise model-free tuning protocols. These are protocols that are able to identify small differences between competing promoter designs and isolate the ones that yield desirable behaviors, e.g., robustness to common perturbations, fast activation times, sufficient temporal spacings, and correct equilibrium concentrations.
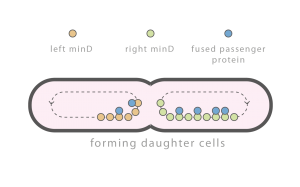
Division Control
Individual cells are constantly subject to perturbations: exogenous perturbations such as temperature fluctuations, brownian motion related perturbations, and perturbations caused by cell division. Cell division related perturbations are primarily caused by random partitioning of molecules between daughter cells and can be difficult to attenuate. Important molecules, e.g., chromosomes, implement complex mechanisms to ensure equal partitioning. Other molecules, e.g., the majority of proteins, are simply partitioned at random. Georgiev Lab is interested in developing simple mechanisms to regulate general protein partitioning.
Time lapse of E. coli with an integrated Min D::GFP fusion protein. Observed spatial-temporal oscillations are critical for correct cell division.
The media player is loading...
Microfluidics
Our Microfluidics Research focuses on the study of microfluidic transport phenomena and the design of microfluidic devices with applications in synthetic biology. Current research projects include design and development of sensitive cell sorting techniques, on-chip cell culture, bio-sensors and cell-to-cell communication analysis.