(Created page with "==Division Control== Accurate protein partitioning through spatial-temporal mechanisms. Individua...") |
|||
| Line 1: | Line 1: | ||
| + | ==Gene Tuning== | ||
| + | |||
| + | ==Modular Transcriptional Riboregulation== | ||
| + | |||
| + | ==Programmable Dynamic Riboregulation== | ||
| + | |||
==Division Control== | ==Division Control== | ||
[[File:wiki_DivisionControl.png|thumb|right|alt=gfp under control of placI.|Accurate protein partitioning through spatial-temporal mechanisms.]] | [[File:wiki_DivisionControl.png|thumb|right|alt=gfp under control of placI.|Accurate protein partitioning through spatial-temporal mechanisms.]] | ||
Revision as of 15:24, 5 March 2015
Contents
Gene Tuning
Modular Transcriptional Riboregulation
Programmable Dynamic Riboregulation
Division Control
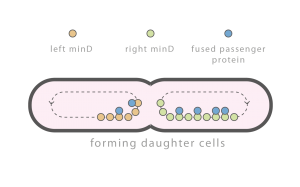
Individual cells are constantly subject to perturbations: exogenous perturbations such as temperature fluctuations, brownian motion related perturbations, and perturbations caused by cell division. Cell division related perturbations are primarily caused by random partitioning of molecules between daughter cells and can be difficult to attenuate. Important molecules, e.g., chromosomes, implement complex mechanisms to ensure equal partitioning. Other molecules, e.g., the majority of proteins, are simply partitioned at random. Georgiev Lab is interested in developing simple mechanisms to regulate general protein partitioning.
Time lapse of E. coli with an integrated Min D::GFP fusion protein. Observed spatial-temporal oscillations are critical for correct cell division.
The media player is loading...