(→Programmable Dynamic Riboregulation) |
(→Modular Transcriptional Riboregulation) |
||
| Line 6: | Line 6: | ||
==Modular Transcriptional Riboregulation== | ==Modular Transcriptional Riboregulation== | ||
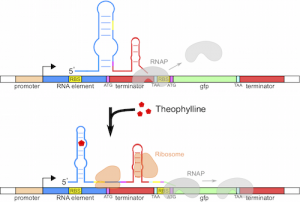
[[File:riboswitch.png|thumb|right|alt=-|Theophylline responsive transcriptional riboregulator.]] | [[File:riboswitch.png|thumb|right|alt=-|Theophylline responsive transcriptional riboregulator.]] | ||
| + | In cooperation with the Universitat Politécnica de Valencia and IBMCP, Spain, we showed that controller inputs based on metabolite measurements can outperform the controller inputs based on enzyme measurements and that transcriptional regulation is advantageous in the regulation of large operons. To address challenges of rational design of transcriptional regulation based on metabolite measurements, we proposed a novel mechanism motivated by easily designable elements from translational regulation. These adaptors use transcriptional-translational coupling in prokaryotes and are potentially scalable to more larger logic circuits. | ||
<br> | <br> | ||
Revision as of 15:42, 5 March 2015
Contents
Gene Tuning
Transcription networks are commonly used in synthetic biology to implement a wide variety of regulatory, logic, and temporal functions. Tuning of transcription networks is commonly achieved by design of the gene promoter regions. Characterized promoter libraries serve well in this design as an initial starting point (Hammer 2006). As with any engineered system, however, precise behavior is attained by in vivo fine tuning. Model based tuning is made difficult by inherent biological model overparametrization (Gutenkunst 2008). Tuning using high throughput assays is also prohibitive both in terms of experimental workload and precision. Georgiev Lab is working on developing precise model-free tuning protocols. These are protocols that are able to identify small differences between competing promoter designs and isolate the ones that yield desirable behaviors, e.g., robustness to common perturbations, fast activation times, sufficient temporal spacings, and correct equilibrium concentrations.
Modular Transcriptional Riboregulation
In cooperation with the Universitat Politécnica de Valencia and IBMCP, Spain, we showed that controller inputs based on metabolite measurements can outperform the controller inputs based on enzyme measurements and that transcriptional regulation is advantageous in the regulation of large operons. To address challenges of rational design of transcriptional regulation based on metabolite measurements, we proposed a novel mechanism motivated by easily designable elements from translational regulation. These adaptors use transcriptional-translational coupling in prokaryotes and are potentially scalable to more larger logic circuits.
Programmable Dynamic Riboregulation
Riboregulators represent an important class of genetic regulatory devices whose behaviour is programmable by specification of the underlying nucleic acid sequence. Both natural and synthetic riboregulators are often modelled as static devices that activate or repress potential RNA binding sites. Individual riboregulators are thereby functionally similar to linear gains found in electrical systems. While in silico design tools have been developed to maximize their in vivo performance, design of dynamic behaviour has yet to be considered. Herein a simple riboregulator design is proposed that appends programmable dynamic behavior to existing riboswitches. Specifically, the resulting device is shown to produce an all-or-nothing response. Simple design rules for this device are derived.
Division Control
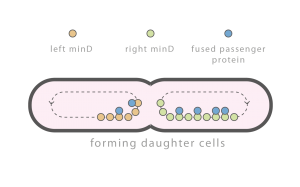
Individual cells are constantly subject to perturbations: exogenous perturbations such as temperature fluctuations, brownian motion related perturbations, and perturbations caused by cell division. Cell division related perturbations are primarily caused by random partitioning of molecules between daughter cells and can be difficult to attenuate. Important molecules, e.g., chromosomes, implement complex mechanisms to ensure equal partitioning. Other molecules, e.g., the majority of proteins, are simply partitioned at random. Georgiev Lab is interested in developing simple mechanisms to regulate general protein partitioning.
Time lapse of E. coli with an integrated Min D::GFP fusion protein. Observed spatial-temporal oscillations are critical for correct cell division.
The media player is loading...